Background
We use the Berlin Harvesting and Indexing Toolkit (B-HIT) to harvest GGBN provider data. The records (or units) can be harvested from providers having either a BioCASe or an IPT installation. For BioCASe providers, the schemata ABCD 2.06, ABCD 2.1, ABCDDNA, ABCDGGBN and ABCDEFG are supported (single records or ABCD Archives). For IPT providers, DarwinCore Archives are supported, including the GGBN extensions. Elements that are indexed are listed at http://wiki.bgbm.org/bhit/index.php/Indexed_fields.
You can either use the full text search on the landing page or the selecting "Search" from the menu
Data Quality and Data Cleaning
During harvesting GGBN provider data are checked and cleaned, if necessary. We keep the original provider data in addition to cleaned versions. Data quality tests are done using B-HIT. Country names are translated in English, ISO codes are compared to the country names, coordinates are validated and checked against both ISO code and country name. In case of incomplete data, the tool is looking into the named areas and localities and tries to extract some information regarding the country or the water body.
Scientific names are parsed using the GBIF Name Parser (http://www.gbif.org/developer/species#parser) and customized regular expressions.
Taxonomic Backbone
After harvesting the scientific names are matched against certain checklists of the GBIF checklist bank. Higher taxa, synonyms and accepted taxa are retrieved, also using the GBIF checklist bank webservice (http://api.gbif.org/v1/species). These checklists include: Prokaryotic Nomenclature Up-to-Date (PNU), Catalogue of Life, NCBI and the GBIF backbone itself.
Search
Search by fields
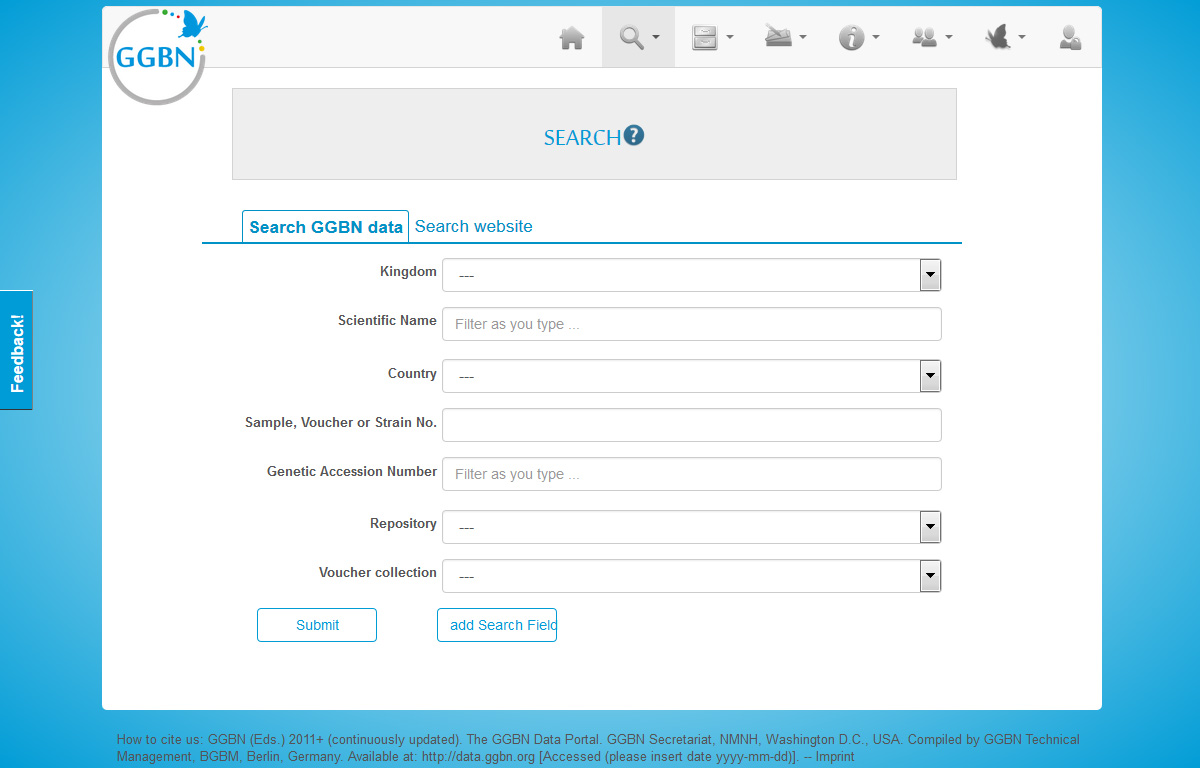
Here you can choose different parameters to filter your results using the facets on the left. The filters/facets are used with AND operator, e.g. materialType=DNA&country=Belgium will search for DNA sample collected in Belgium. Within each facet the OR operator is used, e.g. materialType=DNA&materialType=tissue&country=Belgium will search for DNA OR tissue samples collected in Belgium. This can be extended to as many facets/filters you like.
In addition you can sort the data and preview what kind of material is available by expanding the rows
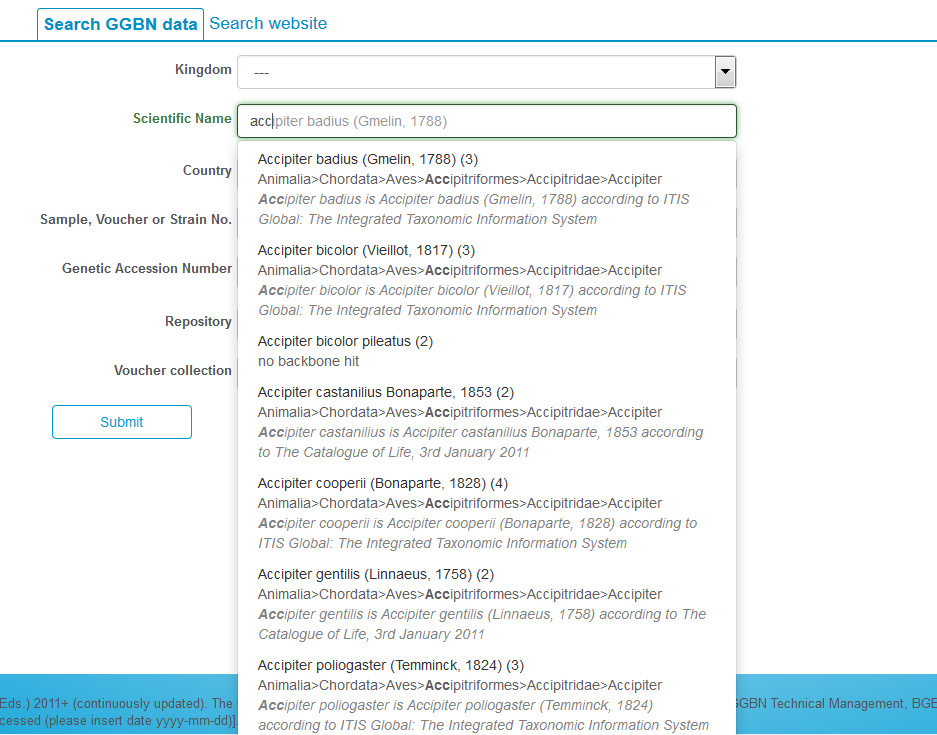
Most of the fields are drop down lists or include suggestion lists to help you. E.g. when typing a name the portal searches for all synonyms and accepted names matching your search term and provides a suggestion list with detailed information about the name found in the GGBN backbone.
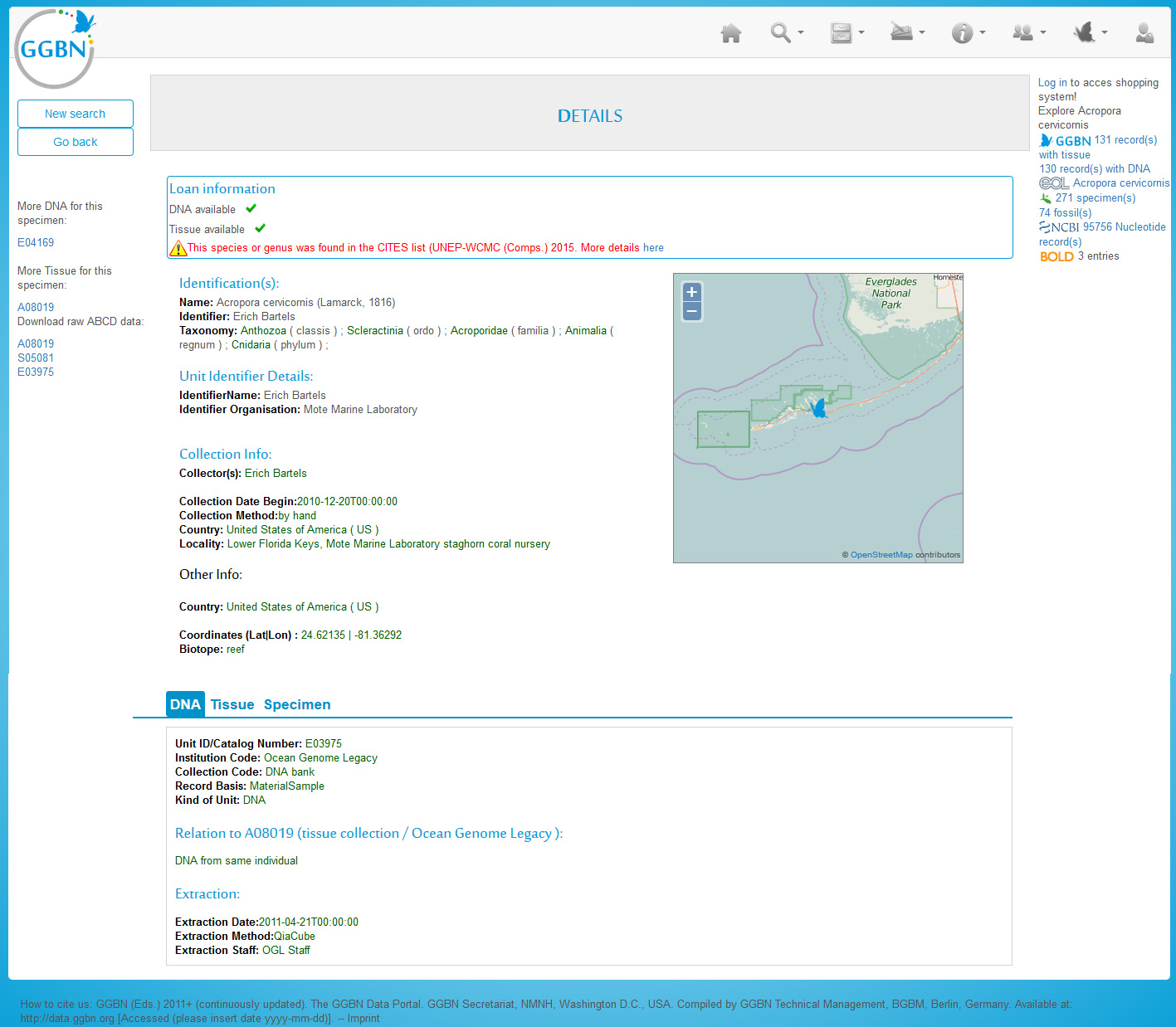
Record detail
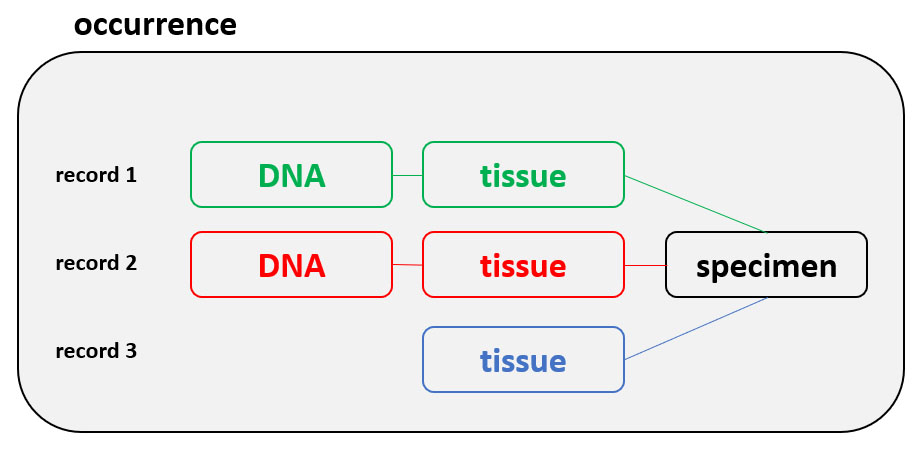
The record details page aggregates data from multiple sources. Please have a look at our definitions on differences between GGBN records, material entities and occurrences. Here you see an example with DNA sample, Tissue sample and Specimen. This is both indicated in the top blue bar (individual tabs for each material entity) and above the title. The data are coming from up to three different datasources, depending on where the samples and data are deposited. Below the map you find information about related records (e.g. another tissue derived from the same specimen) as well as information about loaning availabilities and conditions. Furthermore it is checked whether the taxon is listed on CITES. Left to the map you find collecting information and determination details. In the lower part you'll find with information about the physical samples and where to find them. If you click on the Institution Full Name you'll see the GGBN members page for this institution.
In case sequences or multimedia items are associated to these materials, further tabs will appear. At the very bottom you'll find information about the dataset.